Introduction to Heart Disease Risk Prediction
Heart disease is the leading cause of death globally, requiring an increasing need for early risk prediction vital for preventative healthcare. The objective of this project is to build predictive models for heart disease risk based on clinical data while emphasizing explainability. Traditional black-box models often lack transparency, forming barriers when clinically adopted. Therefore, this project not only implements machine learning and deep learning models for risk prediction but also integrates explainable AI techniques such as SHAP (SHapley Additive exPlanations) to interpret model decisions.
Methodology
Data Collection and Preprocessing
Clinical notes were retrieved from the MIMIC-III database using BigQuery; only summaries categorized under “Discharge summary” were selected. After initial filtering, three numerical features were used: ITEMID, VALUENUM, and rn. Handling of missing values was performed, followed by dataset balancing using SMOTE (Synthetic Minority Oversampling Technique) to address class imbalance. Feature scaling was applied via StandardScaler to ensure uniform input to machine learning models.
Compute Environment
The computation needs of training various models on the MIMIC dataset are non-trivial. After doing multiple experiments on a local MacBook pro and Google Colab environment with the following configurations: Python 3, GPU A100, System RAM: 80 GB, and GPU RAM: 40 GB.
Model Development
Two primary models were trained initially:
- Random Forest Classifier: A tree-based ensemble model known for stability and feature importance extraction.
- Neural Network (Feedforward Sequential Model): A deep learning model consisting of dense layers activated with ReLU and sigmoid functions for binary classification.
The following two models were also utilized in this project: - XGBoost Classifier: An advanced ensemble model optimized for tabular medical datasets. XGBoost provided competitive performance, offering another perspective on predictive accuracy and feature importance in heart disease risk modeling.
- LSTM Sequential Model: An exploratory Long Short-Term Memory (LSTM) model was trained to investigate potential temporal progression patterns in patient data. Due to the static nature of the features, the LSTM model performance was limited.
Both models were evaluated using accuracy, classification reports, and ROC-AUC metrics.
t-SNE Visualization
To explore data structure, t-SNE (t-Distributed Stochastic Neighbor Embedding) was applied on the scaled training data to project high-dimensional patient data into 2D space. Visualization revealed separation between classes after SMOTE balancing.
Explainability with SHAP
To provide insight into model predictions, SHAP explainability was integrated:
- For the Random Forest model, TreeExplainer was used.
- For the Neural Network model, DeepExplainer was applied.
Feature importance plots and force plots were generated to visualize the contributions of individual features to model decisions. SHAP analysis enhanced clinical interpretability by consistently identifying key features like VALUENUM as the primary drivers of heart disease predictions across different models.
SHAP Analysis
- Random Forest SHAP Analysis: Here, VALUENUM emerged as the most influential feature in predicting heart disease, as shown by SHAP analysis.
- Neural Network SHAP Analysis: SHAP explanations similarly highlighted VALUENUM as the primary driver of predictions, ensuring consistency across models.
ROC-AUC Curve Analysis
ROC-AUC curves were plotted to assess model discrimination capability. Comparative analysis was conducted by both Random Forest and Neural Network models.
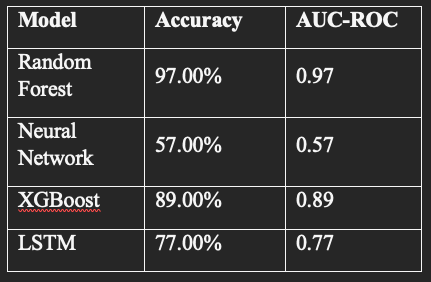
Results: Model Performance Comparison
The performance metrics for all the models used are summarized below:
The Random Forest and XGBoost models demonstrated the strongest discrimination capabilities, with AUC-ROC values close to 0.97. The Neural Network and LSTM models showed lower AUC scores, potentially due to the limited complexity of the network architectures and the nature of static feature inputs. Future improvements could involve fine-tuning hyperparameters or incorporating true sequential clinical data to better leverage the strengths of deep learning models like LSTM.
Conclusion
The study conducted was thus able to successfully demonstrate that combining machine learning and deep learning models with explainable AI techniques enhances heart disease risk prediction. Random Forest and XGBoost models provided strong predictive capabilities, while SHAP explainability added transparency to the model decision-making process. Exploratory models such as the Neural Network and LSTM indicated areas for future research, particularly the integration of true time-series patient data. Future work could extend this by incorporating more complex architectures like TabTransformer or ClinicalBERT, and by focusing on longitudinal modeling with LSTM or GRU based networks.
Related Work
Prior research demonstrated the effectiveness of machine learning in clinical risk prediction. Random Forests have been widely used for tabular health data due to their robustness and interpretability. Deep learning models, particularly feedforward neural networks, have shown promise in capturing complex non-linear patterns. However, the lack of model interpretability remains a critical limitation in healthcare AI. Techniques like SHAP have emerged as strong candidates to bridge this gap, offering local and global feature attribution. This study builds upon these developments by combining predictive modeling with integrated explainability.
References
[1] Breiman, L. (2001). Random forests. Machine Learning
[2] Esteva, A., Kuprel, B., Novoa, R.A., et al. (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature.
[3] Ribeiro, M.T., Singh, S., Guestrin, C. (2016). “Why Should I Trust You?” Explaining the Predictions of Any Classifier. ACM SIGKDD
[4] Lundberg, S.M., Lee, S.I. (2017). A Unified Approach to Interpreting Model Predictions. Advances in Neural Information Processing Systems (NeurIPS)